CLASSICAL METHODS OF TAXONOMY
Aristotle is credited with creating the first classification system more than 2,000 years ago. He classified living things as plant or animal according to their appearance. This system was expanded later by the Romans to be more specific down to the individual organism types, such as a cow or elm tree. As more organisms were identified and classified, more words were needed to separate similar organisms, such as two types of dog. So new organisms were classified by using lengthy descriptors, which eventually became cumbersome to use as more organisms created the need for more words. Carolus Linnaeus, a Swedish naturalist, began the struggle to classify all living things by proposing a binomial, or two-name system. In this model, the genus is the first name, and the species is the second name, with the first letter of the genus name always capitalized. Therefore, each organism is sorted by a genus, species classification, such as Homo sapien for man. Linnaeus also proposed to expand the genus-species nomenclature to include larger units of likeness, for recognizing extended degrees of kinship. Today, the categories are still based on Linnaeus’s work. For example, the taxonomy for humans is the following:
Domain-Eukaryote
Kingdom-Animal
Phylum-Chordate
Order-Primate
Family-Hominidae
Genus-Homo
Subphyla-Vertebrate
Species-Sapien
Class-Mammal
Moving from domain to species, the organisms are more closely related, such that organisms in the same species have a greater degree of kinship than organisms that are similar only at their family level. For instance, both cats and humans are in the class Mammalia, but their pathways separate at that point because cats are in the order Carnivora, whereas humans are Primates. However, a grizzly bear, a black bear, and a polar bear are more closely related because they are in the same family: Ursidae. Further, grizzly bears and black bears are more closely related to each other than to the polar bear because the grizzly and black bear are in the same genus: Ursus, but the polar bear is not. Systematics operates to identify relative kinship and evolutionary intersects among species.
1. Binomial Classification
The binomial classification system proposed by Linnaeus allowed him and others to group organisms together based on common structures, functions, and resulting behaviors, which led to the science of taxonomy, or classification.
Often biologists use a taxonomic key, also known as a dichotomous key, to identify unknown organisms by their physical characteristics. Taxonomic keys work on a base-two premise: The organism either has the characteristic or does not. The resulting answer then directs the biologist to the next set of questions until all the characteristics have been accounted for and the The following Binomial organism is identified. a fictitious key that classification illustration is demonstrates the process.
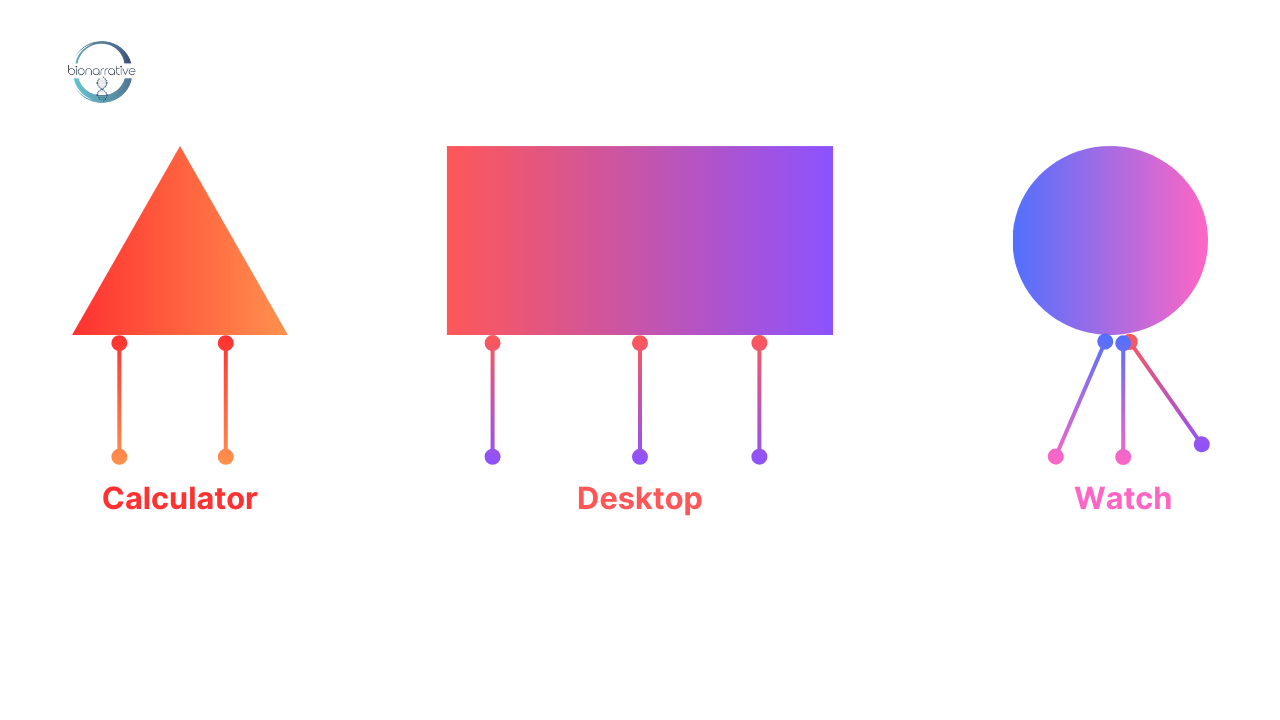
Try using the following taxonomic key to identify which organism is a “laptop.”
| Characteristic | Organism | |
| 1a. | It has two legs. | Calculator |
| 1b. | It has more than two legs. | Go to 2 |
| 2a. | It is shaded. | Go to 3 |
| 2b. | It is not shaded. | Go to 4 |
| 3a. | It has a round head. | Watch |
| 3b. | It does not have a round head. | Laptop |
Backtracking from the dichotomous key, a dumlop is an organism that has more than two legs, is shaded, with a round head. Most dichotomous keys are quite lengthy, to account for all the features that a set of organisms may possess. The broadest keys begin at the kingdom level and proceed to the more specific genus and species levels.
2. Systematics Analysis Techniques
Backmapping the phylogenetic tree for an organism’s evolutionary pathway is the science of systematics. Systematics establishes degrees of kinship, diversity, evolutionary linkages, as well as taxonomic classification for all species.
A common method of establishing kinship is the identification of homologous structures shared by two different species. For instance, a human arm, an alligator foreleg, and a penguin’s flipper are homologous structures not because they look alike or function similarly, but because they both share a common bone structure indicating they all evolved from the same bone feature. The more homologous structures in common, the greater the degree of kinship.
A homologous structure is an anatomical term referring to commonalities in structural characteristics that evolved from the same feature in an ancestor.
Analogous structures are often confused with homologous structures because they appear to be similar in structure and function and resulting behavior. Convergent evolution occurs when dissimilar species living in the same territory and under the same environmental pressures develop analogous or similar structures that function in response to natural selection. As an example, both sharks and whales share common body structures that allow them to swim in the water. However, these structures are analogous, homologous, because they trace their origin to different features: Whales are mammals, and sharks are fish! An embryonic analysis is often used to confirm homologous or analogous structures. Analogous structures do not indicate a degree of kinship.
Molecular Analysis
Anatomical analyses, such a homologous structures, have been the foundation of systematics until the onset of more sophisticated molecular analysis. Because this emerging technology has allowed for the identification and sequencing of proteins and nucleic acids, each organism can be individualized.
Nucleic Acid Analysis
The comparison of DNA nucleotide sequences between two separate specimens is the most accurate method of establishing kinship. Other methods include an analysis of rRNA nucleotide sequences and the extent of hydrogen-bonding similarities between DNA strands.
Amino Acid Analysis
Similarly, a comparison of the sequencing of amino acids in a polypeptide provides the most accurate analysis of protein structures. This analysis of protein backmaps to the DNA that coded for the amino acid sequence. The closer the match between amino acid segments, the more similar the DNA, the closer the phylogenetic relationship.
3. Modern Classification:
Over the years, many models for classifying organisms have been touted as the next best one. Linnaeus’s scheme of classifying everything into two kingdoms was the first real attempt, and it lasted for more than 200 years! However, because living organisms are so diverse, Linnaeus’s classification system is considered inadequate today as a means of accurately representing the degree of relationship between organisms. More advanced thinking created first a five-, then a six-, then a three- kingdom classification system to include all the recent discoveries related to kinship. It is likely that as more discoveries are made and more data processed, the three-kingdom model may be in jeopardy.
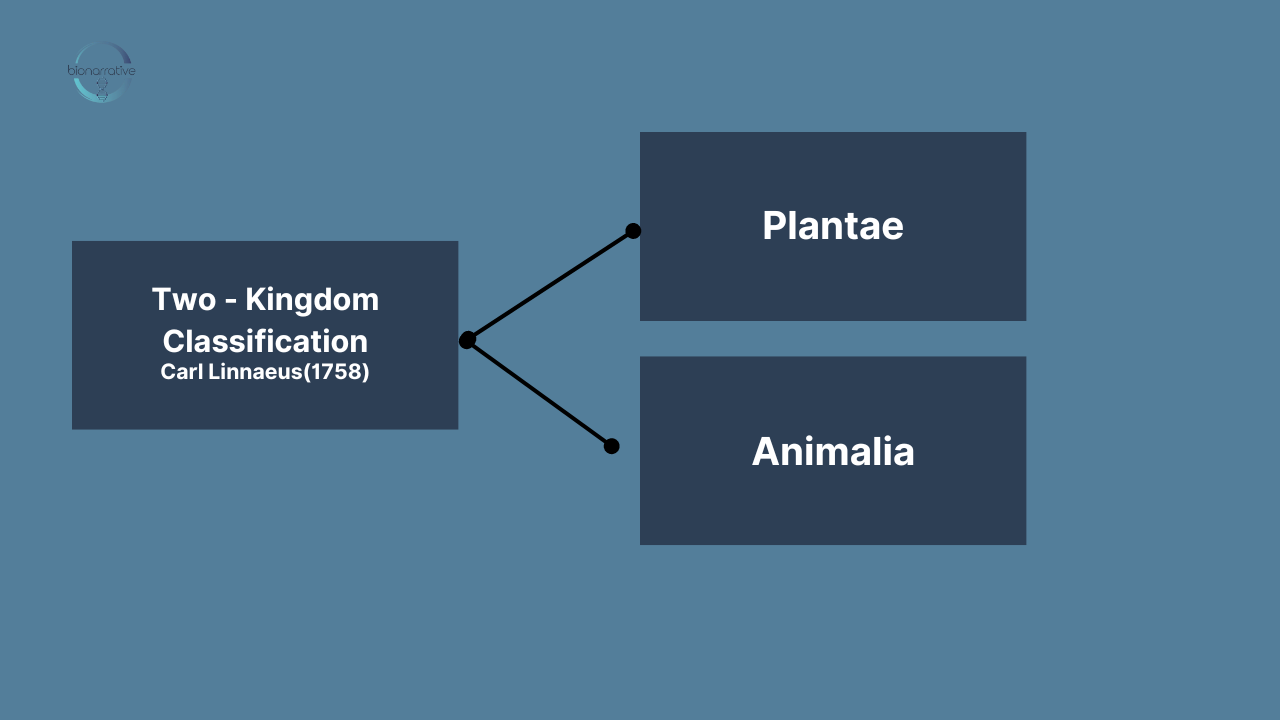
Two-Kingdom Model: Plantae, Animalia
Linnaeus’s model classified everything as either a plant (kingdom Plantae) or animal (kingdom Animalia), which was sufficient for that era. However, even Linnaeus’s followers struggled with the classification of fungi. Most placed them in the plant kingdom because they appeared to have roots and certainly did not belong in the animal kingdom! As additional species were discovered, the two-kingdom classification gave way to other models.
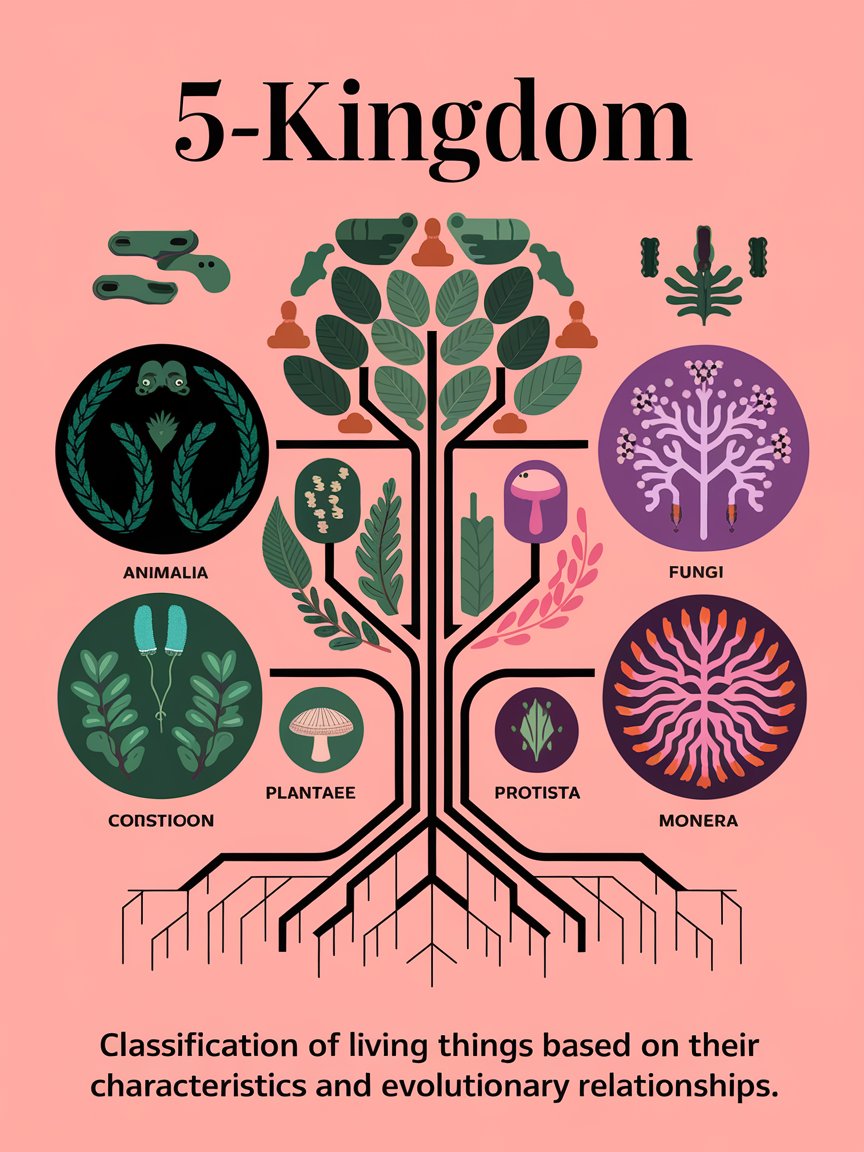
Five-Kingdom Model
American ecologist Robert Whittaker created a five- kingdom model that accounted for prokaryote and eukaryote distinctions. In this schemata, all prokaryotes were placed in the kingdom Monera. The remaining eukaryotes were separated by differences mostly in structure. Plants and animals were easily separated into their own kingdoms. It was decided that because fungi were neither plant nor animal but subsisted by decaying or decomposing once-living matter, they also become their own kingdom. Everything else was clumped into the kingdom Protista. The protists included all eukaryotes that did not clearly fit into the plant, animals, or fungi kingdoms. With the advent of sophisticated collecting and microscopy techniques, the question arose as to where the “new” types of bacteria belong.
Six-Kingdom Model
Linnaeus created his model during the eighteenth century, which proved useful for scientific knowledge a that time. However, recent discoveries of archaebacteria in deep-ocean thermal vents, hot springs in Yellowstone, and brine marine environments justified another look at the five-kingdom model. Analysis of archaebacteria indicates that they are more similar to eukaryotes than their bacterial prokaryotic cousins (see Origin of Prokaryotes and Eukaryotes). The six-kingdom model contains the same original four kingdoms, but splits the Moneran kingdom into kingdom Archaea bacteria to include the newly discovered bacteria type with internal membranes, and kingdom Eubacteria, which contains the more common forms of bacteria. Interestingly, cladistic analysis indicates that organisms in the kingdom Protista are actually more different from each other than originally suspected. In fact, most mammals are more closely related to certain archaebacteria than some protists are related to each other!
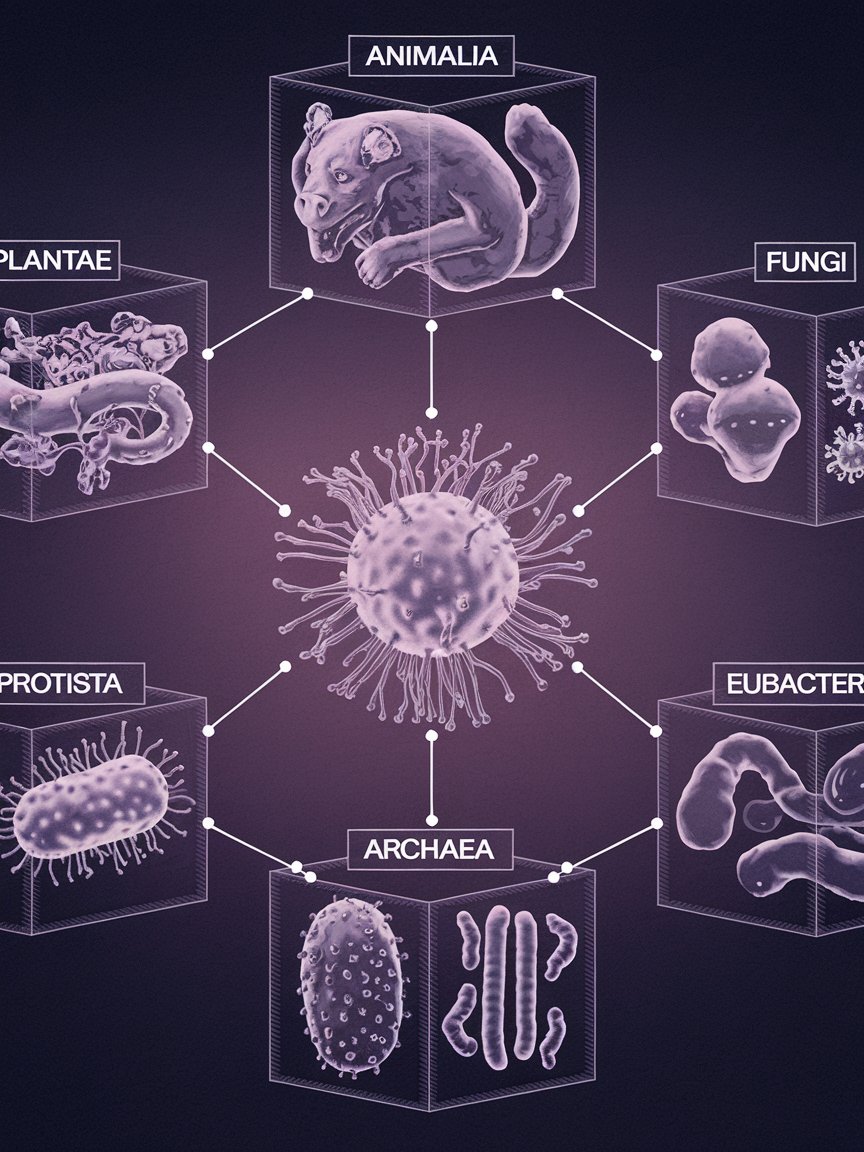
Three-Kingdom Model
The five- and six-kingdom models are based on the scheme of two fundamentally different groups of organisms: prokaryote and eukaryote. They divided and sorted all living things into the various kingdoms based on similarities of evolutionary history. The three- kingdom model makes a fundamental change. Instead of classifying organisms into prokaryote or eukaryote, it sorts them into three categories by splitting the prokaryotes into two kingdoms similar to the six- kingdom model: Archae bacteria and Eubacteria. All the eukaryotes, which is virtually everything else, are clumped into the Eukarya kingdom.
Modern chemical and cellular evidence currently supports the three-kingdom model. It is believed that bacteria were among the first type of life, and the eubacteria separated leaving the archaebacteria and eukarya to continue as one in evolutionary struggles. The fact that modern archaebacteria contain many eukaryotic features indicates their separation occurred after the eubacteria and thereby establishes greater kinship between archaebacteria and eukarya. Archaebacteria are established as a midpoint between eubacteria and eukaraya. Most modern taxonomists favor the three- kingdom model.
2. Alternative methods of classification:
The Linnaean binomial system of classifying animals brought organization from chaos; but recently, with the application of modern technology, new methods have surfaced that yield additional information. Methods of establishing ancestral kinship are helpful in establishing new taxonomic procedures that often relate species in new ways. Although no one method is without drawback, each offers unique insights and information in reference to the organisms in question.
Cladistic Analysis
Cladistic analysis is probably the most widely used alternative method. Cladistic analysis is a means to classify organisms to match their evolutionary history. Common phylogenetic features are used to establish relatedness between organisms with the help of sophisticated computer programs that quickly sort organisms according to shared evolutionary structures.
Cladistic analysis sorts homologous structures into either a primitive character or a derived character. Primitive characters establish the broad classification that generates the basic grouping of organisms. For instance, a cladistic primitive character for plants is the presence of chloroplasts. Those organisms that contain chloroplasts are clumped into the same large grouping.
Derived characters are also homologous structures, but they represent features that have been modified for specific functions. Derived characters are more unique than primitive characters and tend to sort organisms by their presence or absence in the organism. The presence of a derived character or set of derived characters establishes a greater degree of relatedness. The more derived characteristics organisms share, the greater their degree of kinship. For instance, a derived characteristic in plants is the presence of vascular tissue. Advanced plants contain vascular bundles, but simple aquatic plants do not. This relatively simple anatomical feature demonstrates the vast difference between vascular and nonvascular plants.
Review the example that follows to distinguish primitive and derived characters in mammals.
Mammalian primitive characters:
- Mammary glands and hair (for example, humans, dogs, cows, whales)
Derived characters:
- Appendages modified for aquatic movement (for example, whales)
- Appendages with an opposable thumb (for example, humans)
- Appendages designed for running (for example, dogs)
- Appendages designed for grazing on uneven ground and carrying heavy body weight (for example, cows)
After the primitive and derived characters are known, a cladogram can be constructed to show evolutionary linkages between groups of animals. Examine the illustration Simple cladogram.
This cladogram shows the evolutionary relatedness of major plant types by using simple derived characters on the right side in ascending order. Organisms located next to each other horizontally across the top are more related than those not in close proximity. For more specific classifications, such as the kinship between an oak tree and an elm tree, the cladogram would need more specific derived characters.
The cladistic model is somewhat similar to previous models of organization, except for several notable differences. The one most widely reported is the cladistic location of birds on a cladogram in relation to the reptiles. A cladogram relates birds closer to crocodiles and dinosaurs than to snakes or lizards. Interestingly, it is now known that reptiles did not evolve from a common reptilian ancestor, but are more likely descended from several different ancestors, making the reptile classification a conglomeration of animals with similar characteristics but dissimilar backgrounds! The cladogram correctly shows the hereditary linkage between birds and certain reptiles. It clearly is different from classical taxonomic classifications, which place all reptiles in one category (Reptiles) and all birds in another (Aves).
Cladistic analysis is extremely objective: The organism either has the feature or not. This strength is also a weakness. Opponents charge that the technique does not account for the amount or degree by which a feature is present or is utilized. In their opinion, this omission ignores too much relevant data and fails to make an accurate assessment of the difference between groups. For example, the fact that penguins have wings but do not use them to fly would create a cladistic analysis problem.
Phenetics
Phenetics classifications are somewhat similar to evolutionary systematics in that both include all available data regarding the study of organisms and both are antagonistic to the cladistic analysis model. Phenetic classification does not attempt to establish evolutionary linkages but simply the clumping together of organism based on “overall” degrees of similarity. Phenetic classification requires access to the most data. Its overall effectiveness is diminished when the data are incomplete.
Evolutionary Systematics
A contrast to the no-bias approach of the cladistic analysis approach, the evolutionary systematic method deliberately builds in observer judgment. These taxonomists place heavier emphasis on the observed use or nonuse of a structure as well as the way it is used. Judgments are based on direct observation regarding the degree of evolutionary importance a particular feature is to that organism. For instance, in the previous bird and reptile analogy, the evolutionary systematic approach would lend more importance to the presence or absence of feathers than the derived homologous characteristics. So birds would be classified separate from reptiles. Most taxonomists agree that in the absence of data, the cladistic model is superior; with adequate data, the evolutionary systematic model has advantages.